During a pivotal period in genomics history, I found myself at the crossroads of computing and life sciences at Intel, witnessing a breathtaking drop in the cost of sequencing a human genome, from nearly $3 billion to under $1,000. Yet, even as this progress unfolded, we faced a critical challenge: analyzing a single genome could still take weeks. For cancer patients and critically ill newborns, these delays underscored the urgent need to turn scientific possibility into lifesaving reality. The stakes could not have been higher, fueling our sense of purpose.
Understanding the Genomics Analysis Pipeline
The challenge we faced back then wasn't just about speed, but it was about architecting an entire analytical ecosystem. Genomic sequencing analysis operates through three distinct stages: Primary analysis converts raw signals from sequencers into base calls, the order of A, T, C, and G DNA bases. Secondary analysis aligns these sequences to reference genomes and identifies genetic variants; this is the most computationally demanding stage. Tertiary analysis interprets the clinical significance of those variants, matching them against databases of known disease associations and treatment options.
At Intel, our work focused on optimizing across all three stages while building scalable pipeline architectures that could orchestrate hundreds of concurrent analyses and integrate results into clinical systems.
Vision and Strategic Partnerships (2014-2016)
At Intel1, we set an audacious vision: deliver complete diagnosis and genetic analysis within 24 hours,"All in One Day."2 Collaborating with trailblazing partners such as Ayasdi3, we achieved remarkable breakthroughs, including a 400% performance improvement for the Ayasdi Cure application designed to help pharmaceutical and biotech companies accelerate the drug discovery process by 50%. Working with GenomeNext, AWS, and Nationwide Children's Hospital4, we proved it was possible to analyze 1,000 complete genomes in a single day. Our alliance with Edico Genome5 propelled us even further, enabling us to analyze a whole genome 70x faster and complete the analysis in 20 minutes. Jointly with Janssen R&D (a division of Janssen Pharmaceutica) and IMEC (Interuniversity Micro-Electronic Center, Belgium), we built elPrep for preparing SAM/BAM/CRAM files for variant calling in next-generation sequence analysis workflow6. Each milestone brought us closer to transforming patient care and saving lives.
We established the Collaborative Cancer Cloud7,8, a federated architecture (shared but decentralized) that allows institutions to share insights while keeping raw data private. In 2016, our $25 million partnership with the Broad Institute9 produced Cromwell, a workflow engine for genomic pipelines at cloud scale, and optimized GATK (Genome Analysis ToolKit), reducing the time for 20,000-sample genotyping by 90%.
A Decade of Transformation
The past decade has been one of extraordinary transformation. Sequencing costs have dropped below $1,000 per genome, once-unimaginable FDA-approved tests are now reality, and over 26 million Americans10 have unlocked their genetic stories. The computational tools we pioneered - cloud-based pipelines, federated architectures, and AI-powered interpretation - are now standard, shaping the future of precision health. This journey has inspired not just innovation, but a profound shift in what is possible for patients everywhere.
Yet challenges persisted. Regulatory evolution proved complex. Insurance coverage emerged as the most significant barrier, even after Medicare coverage determinations, and claim denial rates increased. Costs ranging from $60011 to over $3,000 creates a substantial patient burden. The "cycle of inaction" persists: physicians are reluctant to order tests without actionable treatments, and payers are unwilling to cover tests not yet the standard of care.
The computational infrastructure succeeded brilliantly. Yet systemic adoption lagged behind technical capability.
The Multiomics Revolution
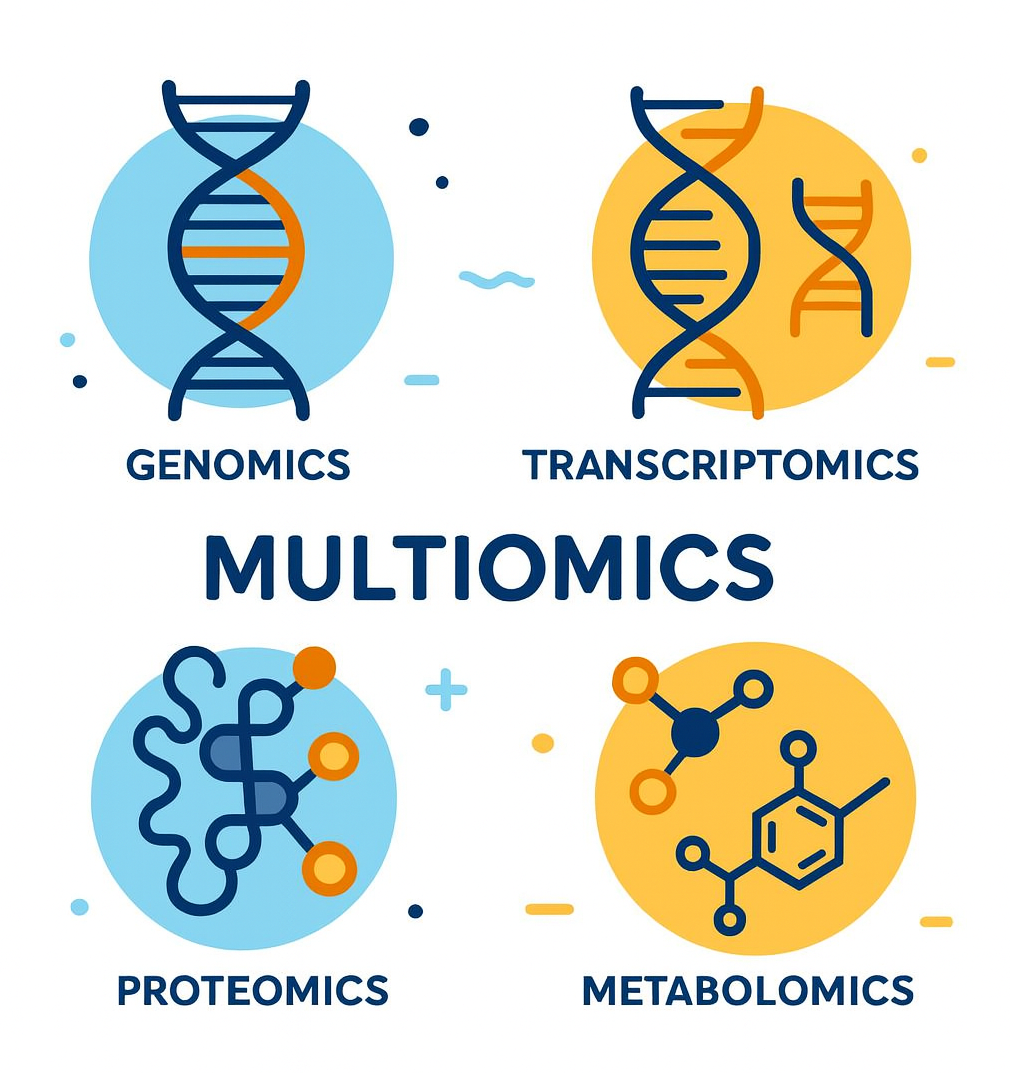
While focusing on genomics, a broader revolution emerged. Genomics shows predisposition, what might happen from inherited DNA, but the genome is static and doesn't show current body states. This realization fueled multiomics, which integrates more biological data layers.
Transcriptomics measures which genes are actively being expressed (i.e., turned on) and which portions of the genetic blueprint are actually being read and translated into action. Two patients with identical genetic variants may have vastly different health outcomes based on which genes are turned "on" or "off." In cancer, transcriptomic analysis reveals which pathways are actively driving tumor growth, enabling more targeted therapy selection.
Proteomics analyzes the actual proteins present in cells and blood. Proteins are the molecular machines that perform biological functions. While genomics tells us which proteins could be made, transcriptomics shows which instructions are being read, and proteomics reveals which proteins are actually present and in what quantities. Protein biomarkers can indicate disease presence, progression, and treatment response far more directly than genetic variants alone.
Metabolomics measures small molecules like sugars, lipids, and amino acids, which are the end products of cellular processes. These metabolites provide real-time snapshots of metabolic activity. Metabolomic signatures, or unique patterns of these small molecules, can identify risk for diabetes, cardiovascular disease, and even mental health conditions by revealing how the body processes nutrients and generates energy.
Epigenomics studies chemical changes (modifications) to DNA that control when and where genes are turned on or off, without altering the DNA sequence itself. These changes are caused by the environment, lifestyle, and aging. Epigenetic markers can indicate a person’s biological age (rather than their actual age), predict disease risk, and reveal how quickly someone is aging. Unlike the underlying DNA sequence, which doesn’t change, epigenetic patterns shift over time, giving insight into dynamic health processes.
The Microbiome represents the collective genomes of trillions of microbes inhabiting our bodies, essentially a second genome with 100 times more genes than our own. The microbiome affects digestion, immune function, mental health, and disease risk. Microbiome composition changes with diet, medication, and environment, making it both a diagnostic tool and therapeutic target.
The Power of Integration
The true revolution comes from integrating these data layers. Consider type 1 diabetes: genomics identifies inherited susceptibility, transcriptomics reveals immune pathway activation, proteomics detects inflammatory markers, metabolomics shows beta cell dysfunction, and microbiomics identifies viral infections or dysbiosis that may trigger disease. Studies combining these approaches can predict which newly diagnosed patients will experience rapid beta cell loss, enabling earlier, more aggressive intervention.
In cancer care, multiomics integration has become transformative. Genomic analysis identifies targetable mutations, transcriptomics reveals active pathways, proteomics confirms protein expression levels that predict drug response, and metabolomics indicates tumor metabolic patterns. This integrated view enables selection of therapies most likely to work for each patient's specific tumor biology.
For complex diseases such as Alzheimer's and cardiovascular disease, where multiple genes and environmental factors interact, multiomics provides the comprehensive view necessary for risk prediction and prevention. Polygenic risk scores (PRS) from genomics, combined with metabolomic markers, proteomic biomarkers, and epigenetic age, create far more accurate predictions than any single data type alone.
Returning to the Mission: Bioscope.ai
After nearly a decade away from the field, I am invigorated to return through Bioscope.ai, a venture born from the enduring conviction that the challenge of translating genomics into real-world, actionable insights can and must be solved. The computational infrastructure we built proved large-scale genomics is feasible. Now the imperative is clear: ensure that these advances empower physicians with timely insights and make precision health an affordable reality for every patient.
Bioscope.ai represents the convergence of everything we had envisioned: whole-genome sequencing integrated with electronic medical records (EMRs), microbiome analysis, and AI to deliver comprehensive clinical decision support. The platform will expand to incorporate additional omics layers, such as epigenomics and proteomics, as these technologies mature.
From Data to Clinical Action
Our earlier work focused on making genomic analysis faster and cheaper. Bioscope.ai solves the challenge of tertiary analysis by combining genomic data with continuously updated medical knowledge bases. The platform doesn't just identify variants; it contextualizes them against a patient's complete medical picture, environmental factors, and microbiome, with plans to integrate transcriptomic, proteomic, metabolomic, and epigenomic data.
The synoptic dashboard synthesizes genomic predispositions with real-world clinical data across 16 health dimensions. This bridges the gap between genomic variants and clinical decisions. It can diagnose vague complaints by combining genomic variants, microbiome analysis, and clinical symptoms to identify root causes that are otherwise difficult to diagnose.
Most compelling is the AI-powered clinical colleague, offering pre-built prompts: "What's new?" "What recommendations?" "Create a personalized longevity plan." This addresses physician time constraints and knowledge gaps we identified through our research.
The Promise Realized
The decade since my earlier work has vindicated the vision while revealing its limitations. My old team succeeded in breaking computational barriers and building scalable architectures. Yet computational capability alone proved insufficient.
Bioscope.ai aims to solve the entire problem, not just analysis, but meaningful interpretation integrated into clinical workflows at a price point that makes adoption feasible. The platform continually reinterprets genomic data as new discoveries emerge, unlike static genetic tests performed once.
Now, as I return to the world of genomics, my optimism has never been greater. We stand on the threshold of a new era, equipped with the technology, tools, and insights to realize the true promise of precision health. By bridging clinical and biological data, we can finally turn hope into reality for countless patients, fulfilling a mission that once seemed impossible.
Systemic barriers such as regulation, reimbursement, and clinical adoption remain. But by addressing usability and demonstrating clear clinical value through integrated multiomics analysis, we can break through the "cycle of inaction."
.svg)







.png)